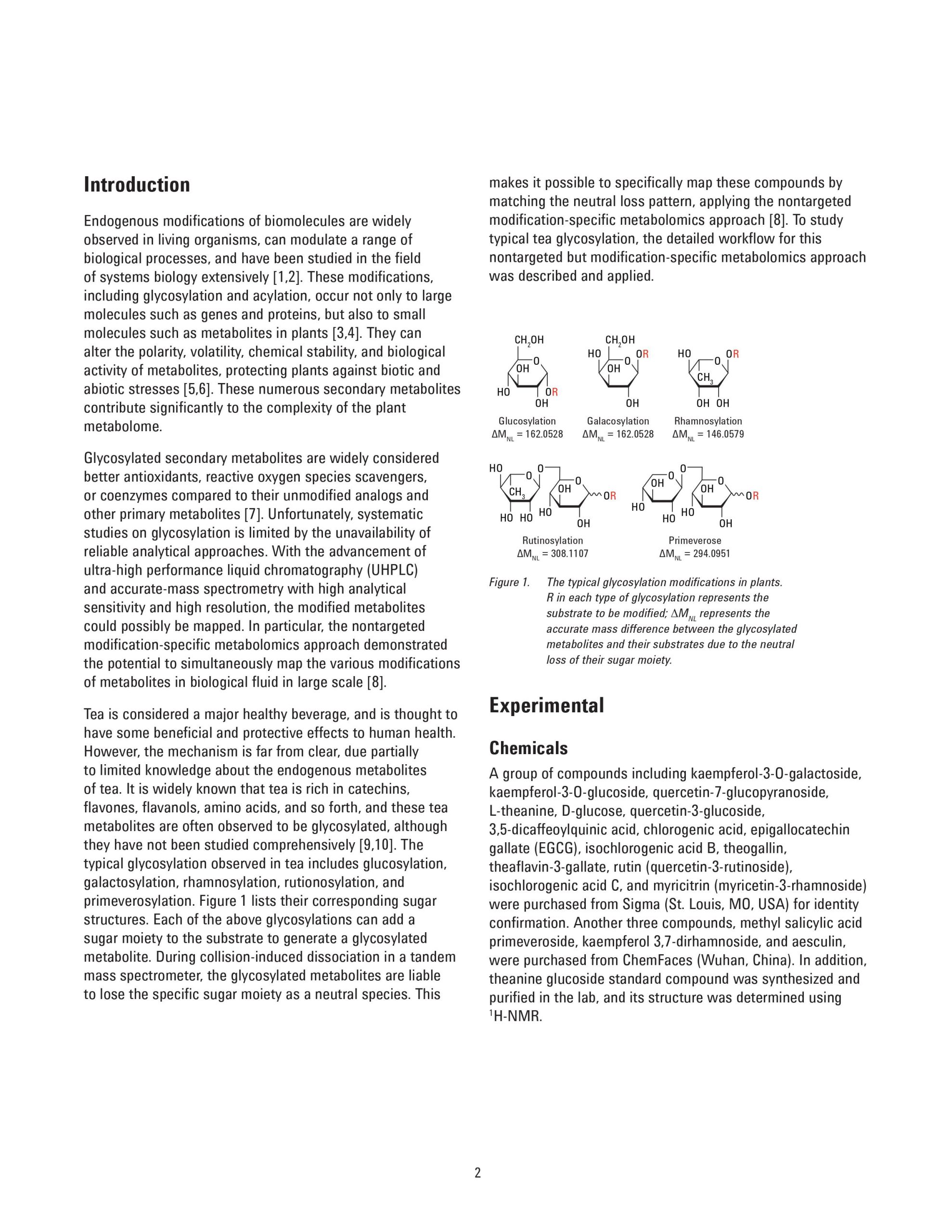
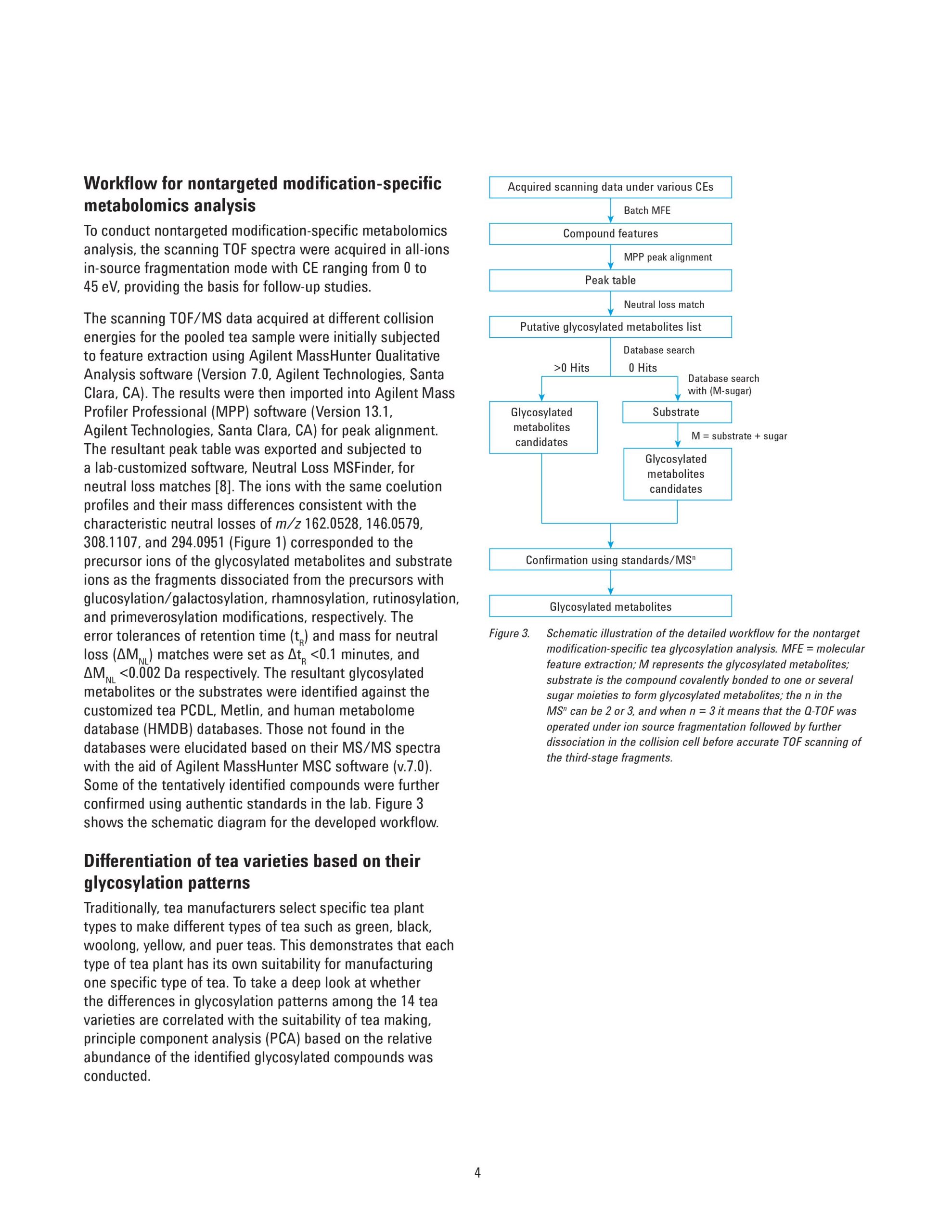
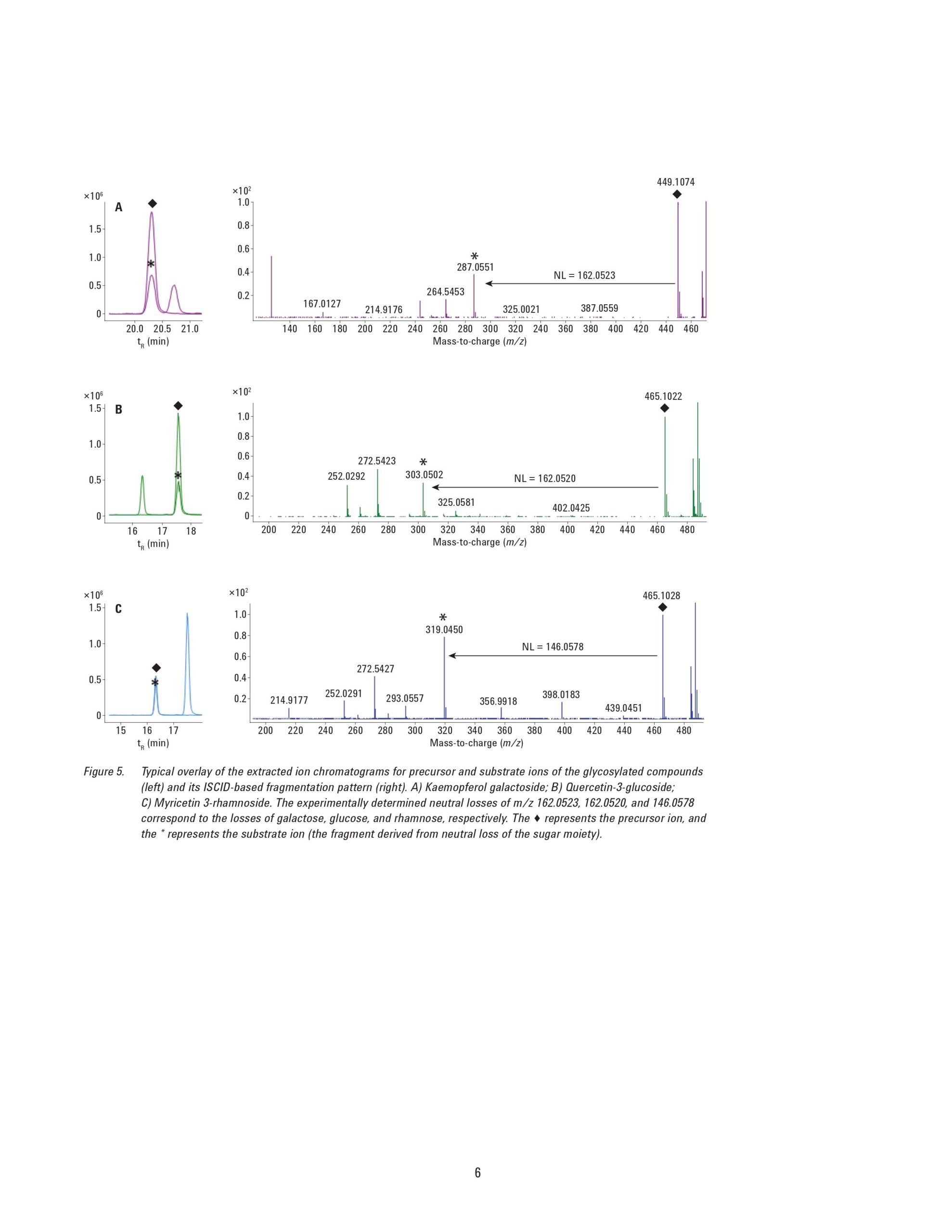
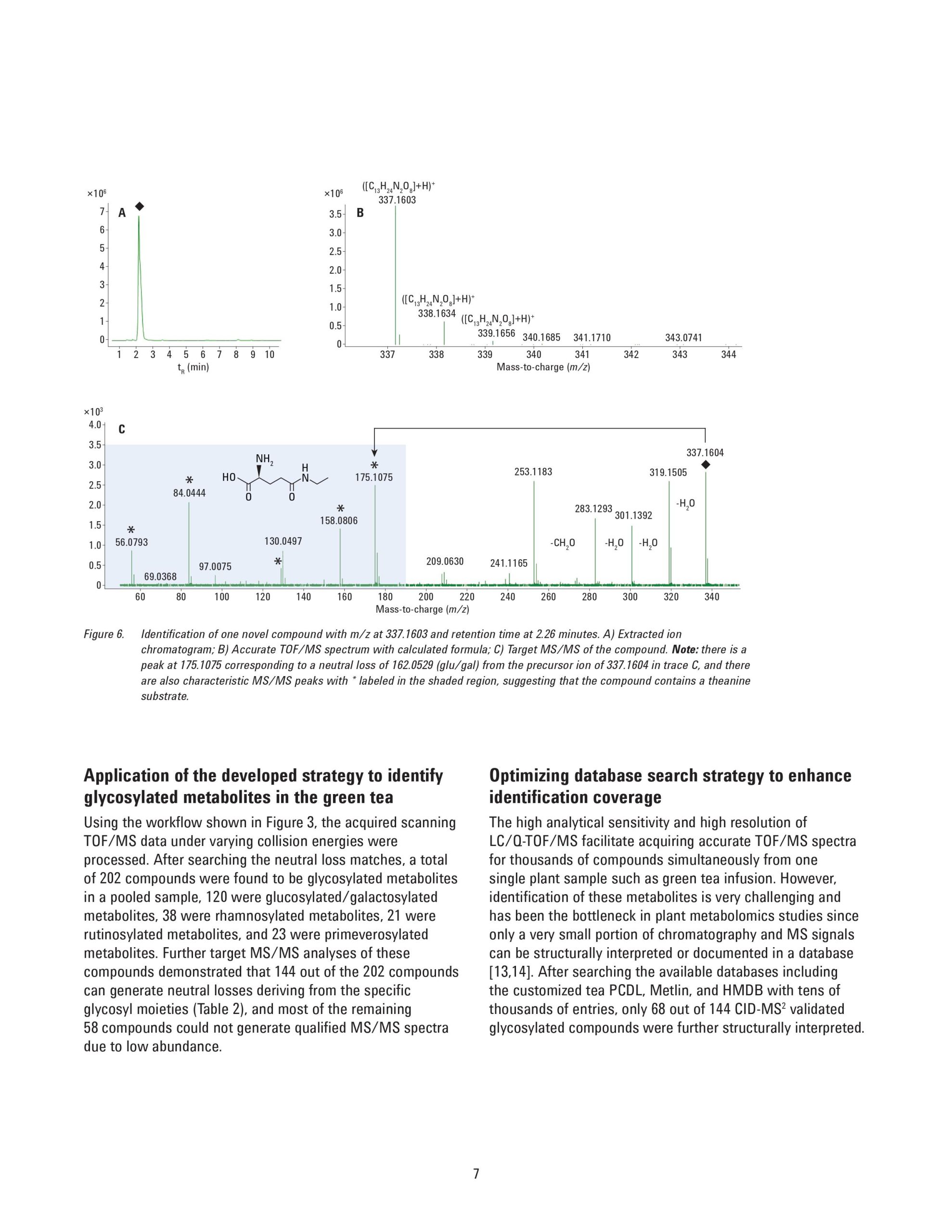
Glycosylation is widely involved in a series of biological events in plants and are believed to be beneficial to human health, but studies on this biological process are limited mainly due to the unavailability of a powerful analytical approach to identify the abundant and diverse glycosylated metabolites in plants. This application note describes a nontarget modification-specific metabolomics approach to study glycosylation in tea (Camellia Sinensis L.), which was recently reported by Dai, et al. [1]. Agilent ultra-high performance liquid chromatography (UHPLC) combined with high-resolution quadrupole time-of-flight mass spectrometry (Q-TOF/MS) was applied to profile the green teas under all-ions in-source fragmentation mode. The acquired data were subjected to a lab-customized workflow to selectively extract glycosylation-related compound features based on the characteristic neutral loss patterns from the specific glycosylation modifications. Further identification of these compounds was based on searching the database directly against the glycosylated metabolites or the corresponding substrates. With this strategy, 202 glycosylated metabolites including glucosylation/galacosylation, rhamnosylation, rutinosylation, and primeverose were detected simultaneously, among which 68 glycosylated metabolites were putatively identified in the green tea infusion. An additional 44 novel glycosylated metabolites were tentatively elucidated based on their MS/MS spectra. This approach allows the user to profile, discover, and identify novel glycosylated metabolites in plant samples.







 Tiếng Việt
Tiếng Việt
